머신러닝의 많은 문제는 train sample 이 수천에서 수백만개의 특성을 가지고 있다는 것입니다. 사람은 3차원 공간에서 살고 있기에 우리가 보고 느낄 수 있는 것들은 1,2,3차원 입니다. 하지만 머신러닝에서는 다루는 차원의 수가 정말 큽니다. 따라서 우리는 매우 간단한 형태이더라도 차원의 수가 높아지면 이해할 수 없어집니다. 그리고 많은 특성은 triain 시간을 느리게 할 뿐만아니라, 좋은 솔루션을 찾기 힘들게 합니다. 우리는 종종 이런 문제를 차원의 저주(CURSE OF DIMENSIONALITY)라고 부릅니다.
하지만 특성 수를 크게 줄여 고차원 공간을 우리가 이해할 수 있는 저차원으로 변환하는 기술을 연구하였습니다. 이를 dimensionality reduction이라고 합니다. 예를 들어 MNIST 이미지를 보면, 이미지 경계에 있는 픽셀은 흰색이므로 이런 픽셀을 완전히 제거해도 많은 정보를 잃지는 않습니다.

차원을 축소시켜 일부 정보를 없앤다면 훈련속도가 빨라지는 것 외에 데이터 시각화에도 아주 유용하게 쓰일 수 있습니다. 차원 수를 둘이나 셋으로 줄이면 하나의 압축된 그래프로 그릴 수 있고 군집같은 시각적인 패턴을 감지해 인사이트를 얻을 수 있습니다.
차원 축소 기법은 투영projection 과 매니폴드 학습manifold learning, 가장 인기있는 PCA가 있습니다.
PCA(주성분 분석)
가장 인기 있는 차원 축소 알고리즘입니다.
사이킷런의 PCA - SVD 분해 방법을 사용하여 구현합니다.
MNIST는 가장 간단한 이미지 데이터셋입니다. 이것은 28 x 28 pixel 의 숫자 이미지로 구성되어있습니다. 실제로 데이터를 불러와서 확인해 보겠습니다.
먼저 불러온 데이터를 이미지와 그 숫자가 무엇인지를 알려주는 label로 나눠주겠습니다. 각각의 이미지는 28 x 28 pixel들을 가지고 있기 때문에 우리는 28×28=784 차원의 벡터를 가지게 됩니다. 하지만 784차원의 공간에서 우리의 MNIST 가 차지하는 공간은 매우 작을 것입니다. 784 차원에는 매우매우 많은 벡터들이 존재합니다.
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.datasets import fetch_openml
mnist = fetch_openml('mnist_784', cache=False)
X = mnist.data.astype('float32').to_numpy()
y = mnist.target.astype('int64').to_numpy()PCA 는 Principal Components Analysis 의 약자로, 데이터가 가장 흩어져있는 축을 찾아서 그곳으로 사영해서 원하는 차원 개수만큼 줄이는 방법입니다. 데이터가 가장 흩어져있는 축이라는 말은 가장 variance 가 커지게 하는 축이라는 말과 같습니다.
PCA를 scikit-learn 패키지를 활용해서 나타내면서 이해해보도록 하겠습니다. 42000개의 데이터는 개수가 너무 많기 때문에 개수를 좀 줄여서 15000개를 가지고 진행하겠습니다.
labels = y[:15000]
data = X[:15000]
print("the shape of sample data = ", data.shape)그리고 feature의 개수가 매우 많이 때문에 정규화를 시켜줍니다. Sklearn 패키지 안의 StandardScaler 함수를 통해서 z-score 정규화를 시켜주겠습니다
from sklearn.preprocessing import StandardScaler
standardized_data = StandardScaler().fit_transform(data)
print(standardized_data.shape)
sample_data = standardized_data2차원으로 축소를 할 것이기 때문에 number of components를 2로 해주겠습니다.
from sklearn import decomposition
pca = decomposition.PCA()
# configuring the parameteres
# the number of components = 2
pca.n_components = 2
pca_data = pca.fit_transform(sample_data)
# pca_reduced will contain the 2-d projects of simple data
print("shape of pca_reduced.shape = ", pca_data.shape)원래 우리가 가지고 있던 데이터는 784차원이었는데 PCA를 통해서 2로 줄어든 것을 확인할 수 있습니다.
이제 이것을 시각화해서 보도록 하겠습니다. 라벨마다 색을 부여해서 시각화하겠습니다.
# attaching the label for each 2-d data point
pca_data = np.vstack((pca_data.T, labels)).T
import seaborn as sn
# creating a new data fram which help us in ploting the result data
pca_df = pd.DataFrame(data=pca_data, columns=("1st_principal", "2nd_principal", "label"))
sn.FacetGrid(pca_df, hue="label", size=6).map(plt.scatter, '1st_principal', '2nd_principal').add_legend()
plt.show()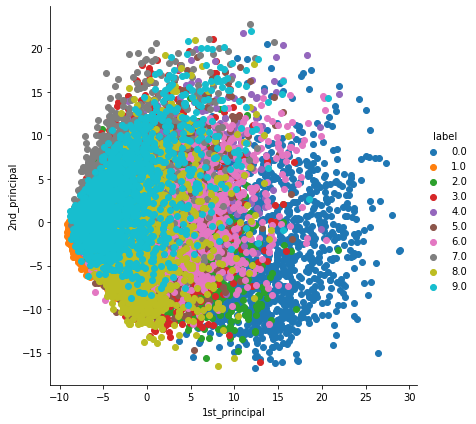
이렇게 우리의 MNIST 데이터셋을 2D로 차원을 축소해서 시각화를 해보았습니다. 비슷한 라벨의 이미지들끼리 모여있는 것을 보아 잘 축소된 것을 알 수 있습니다.
'머신러닝' 카테고리의 다른 글
| 비지도학습 - 군집, k-means, 실루엣점수 사이킷런 (0) | 2022.12.07 |
|---|---|
| 머신러닝 분류 성능 측정 (0) | 2022.12.06 |
| 다중 로지스틱 회귀 (소프트맥스 회귀) (0) | 2022.12.05 |
| 로지스틱 회귀 sklearn logistic regression iris python (0) | 2022.12.04 |
| [머신러닝4] Logistic Regression 로지스틱 회귀 pyhton (0) | 2022.11.30 |